Monitoring SARS-CoV-2 variants alterations in Nice neighborhoods by wastewater nanopore sequencing - The Lancet Regional Health – Europe

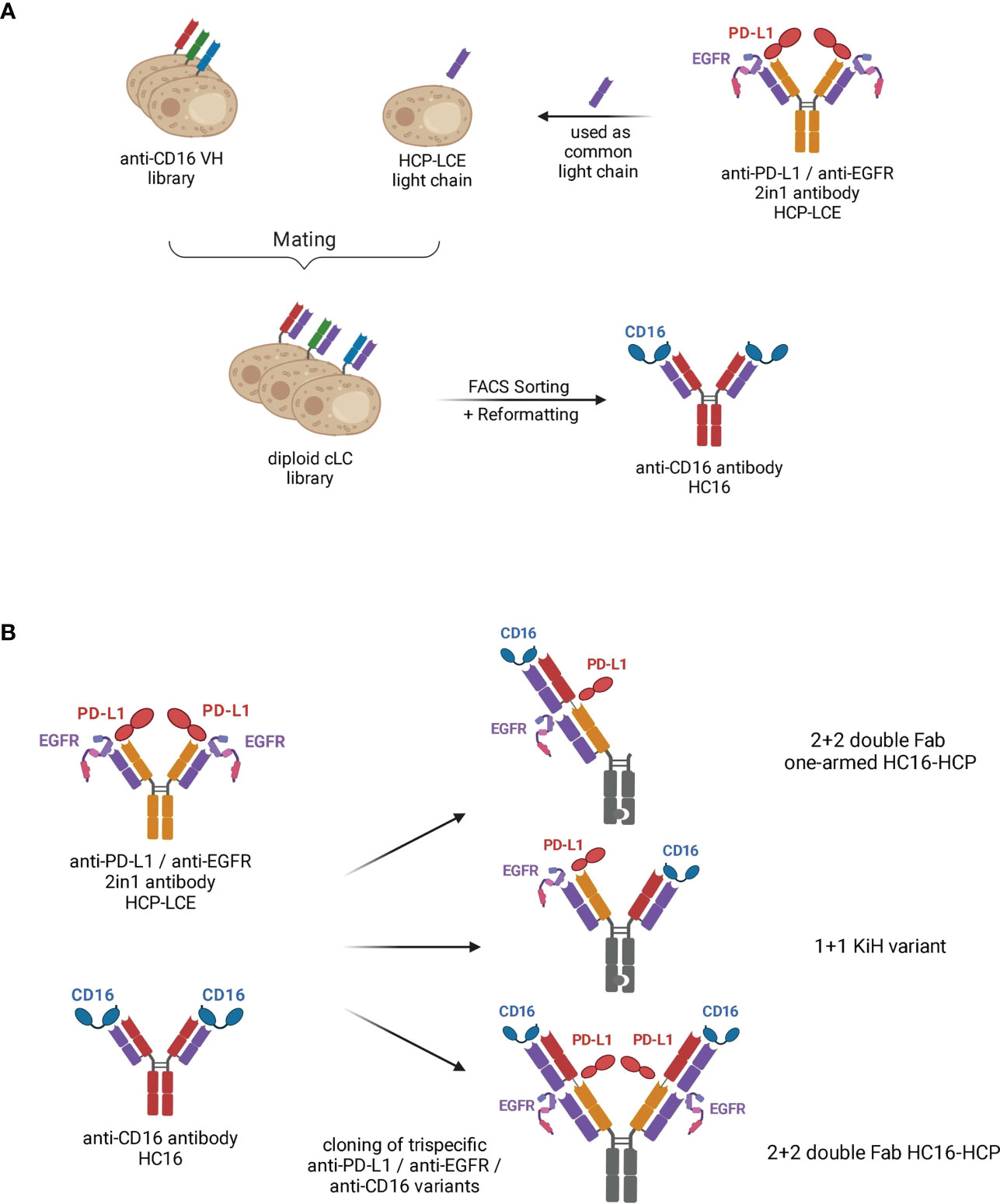
Human inhalable antibody fragments neutralizing SARS-CoV-2 variants for COVID-19 therapy: Molecular Therapy

Protocol to isolate and assess spike protein cleavage in SARS-CoV-2 variants obtained from clinical COVID-19 samples - ScienceDirect
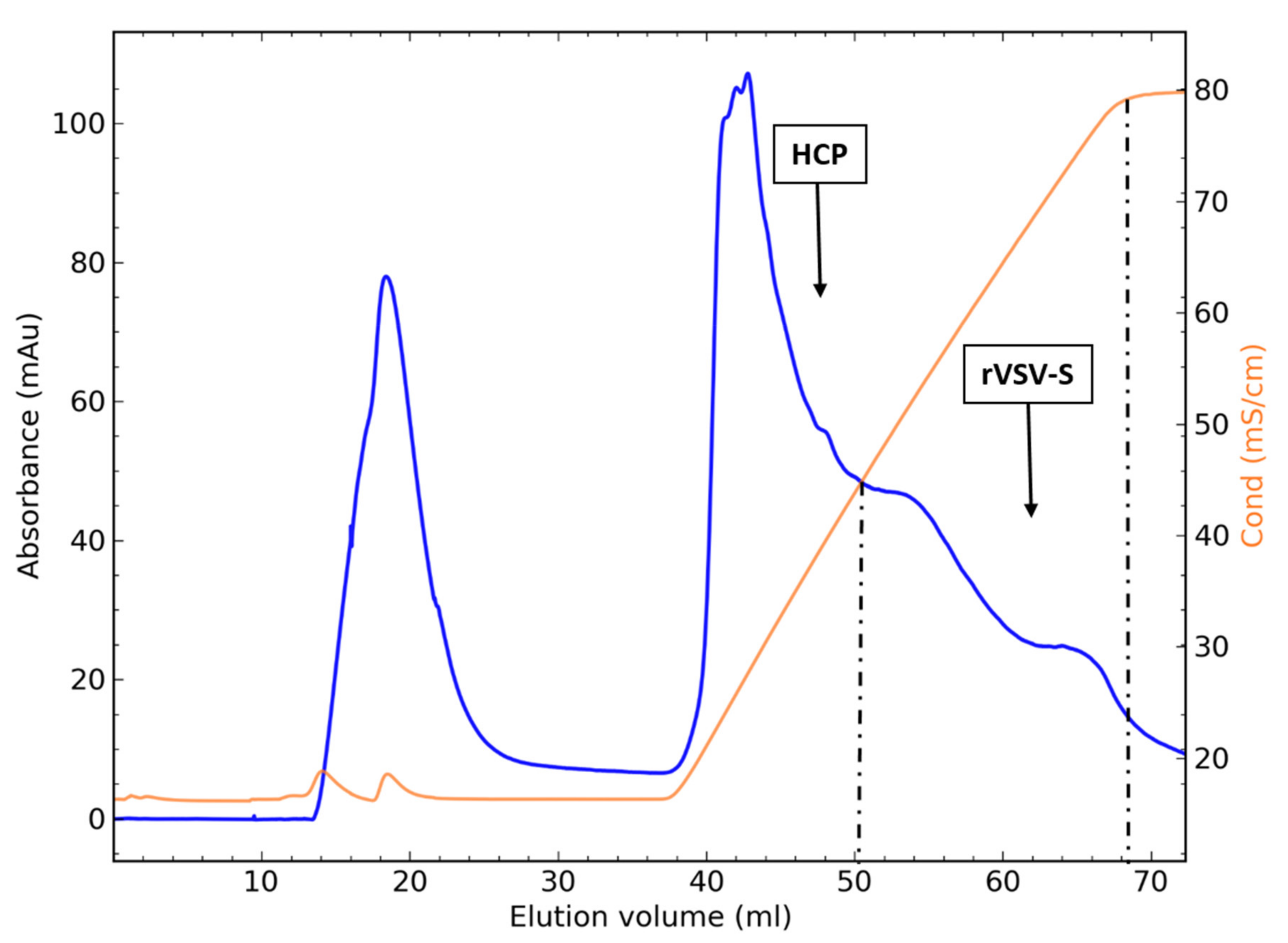
BioTech | Free Full-Text | Highly Efficient Purification of Recombinant VSV-∆G-Spike Vaccine against SARS-CoV-2 by Flow-Through Chromatography

Optimized expression and purification of a soluble BMP2 variant based on in-silico design - ScienceDirect
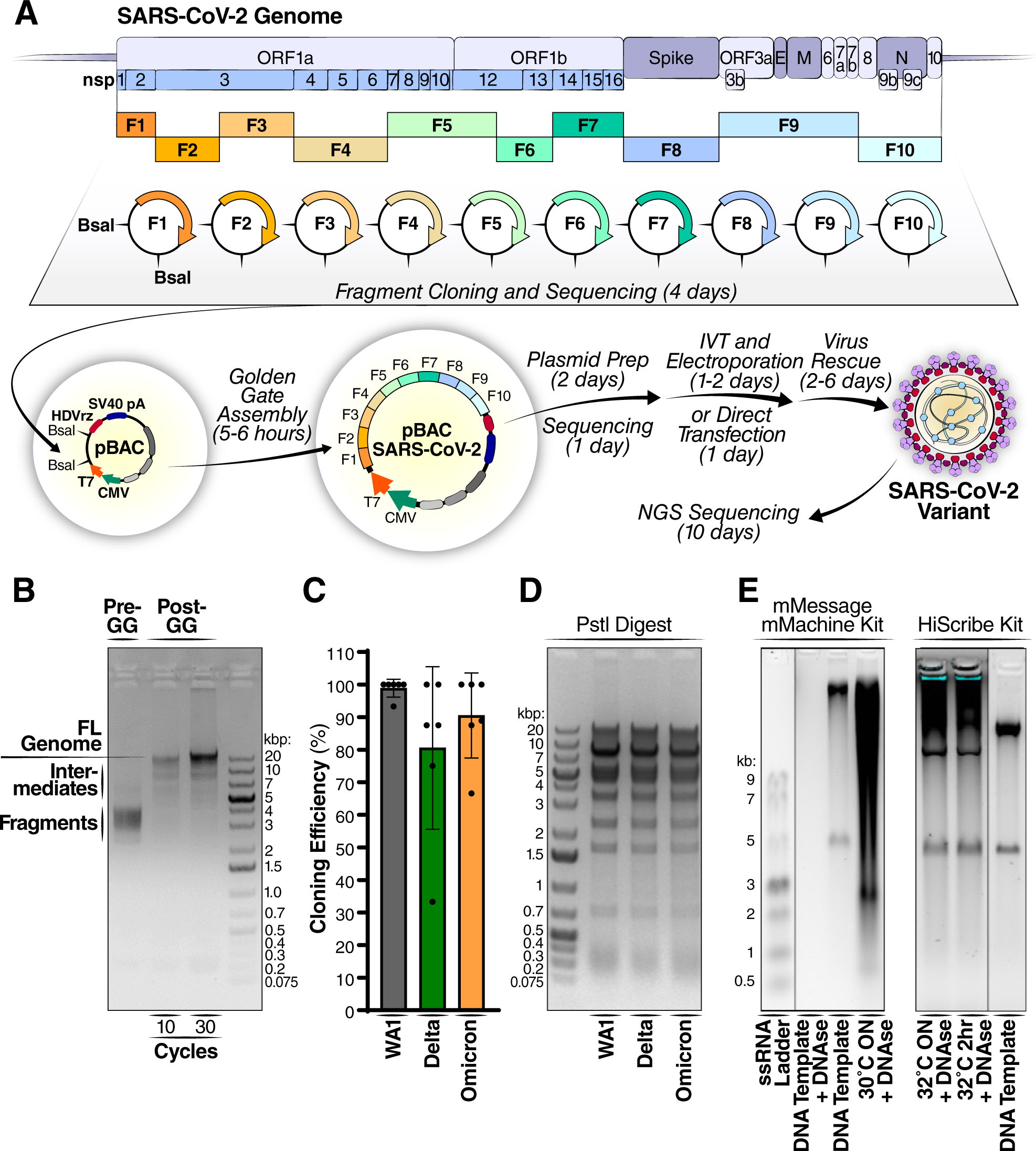
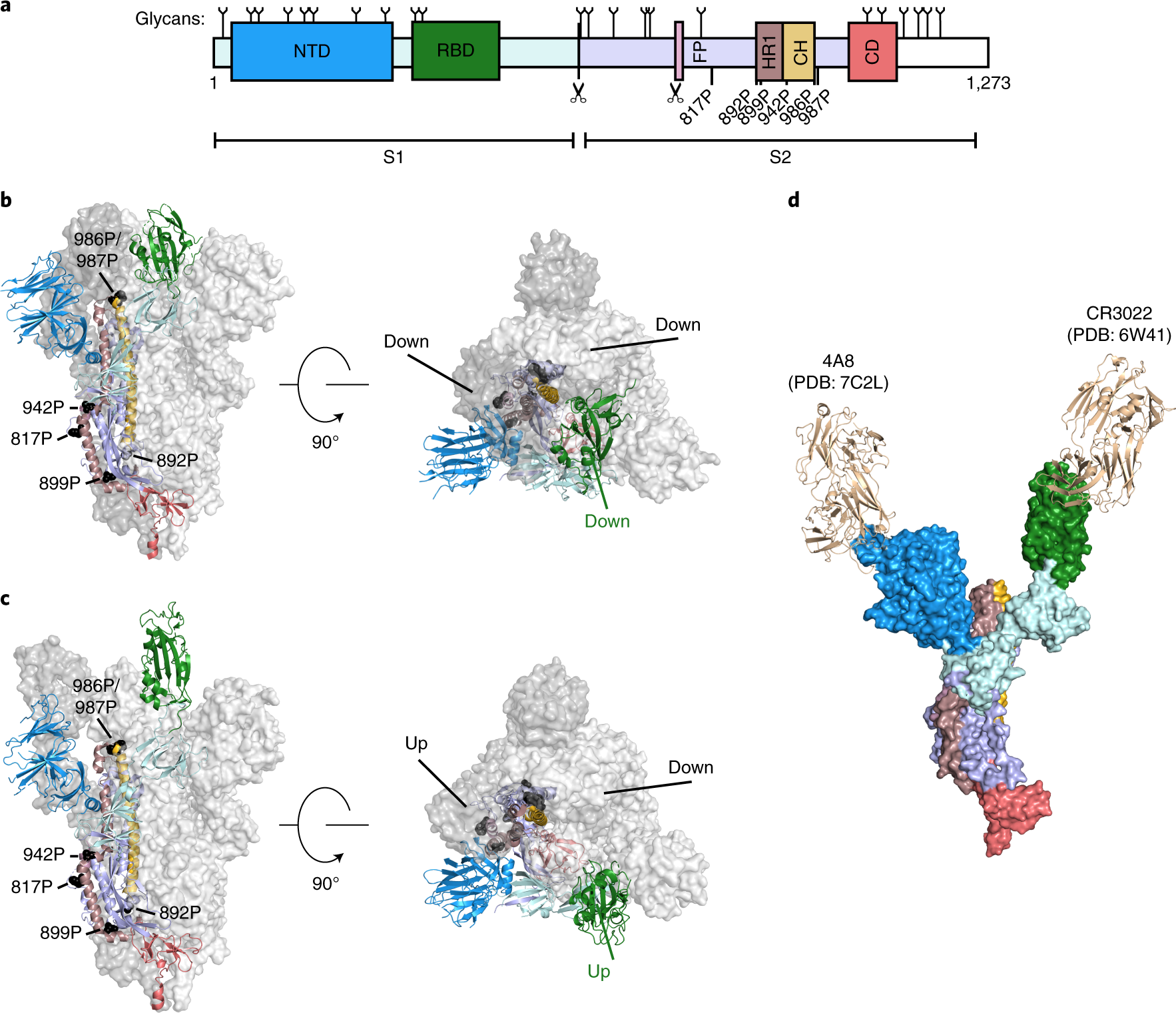
Rapid assembly of SARS-CoV-2 genomes reveals attenuation of the Omicron BA.1 variant through NSP6 | Nature Communications
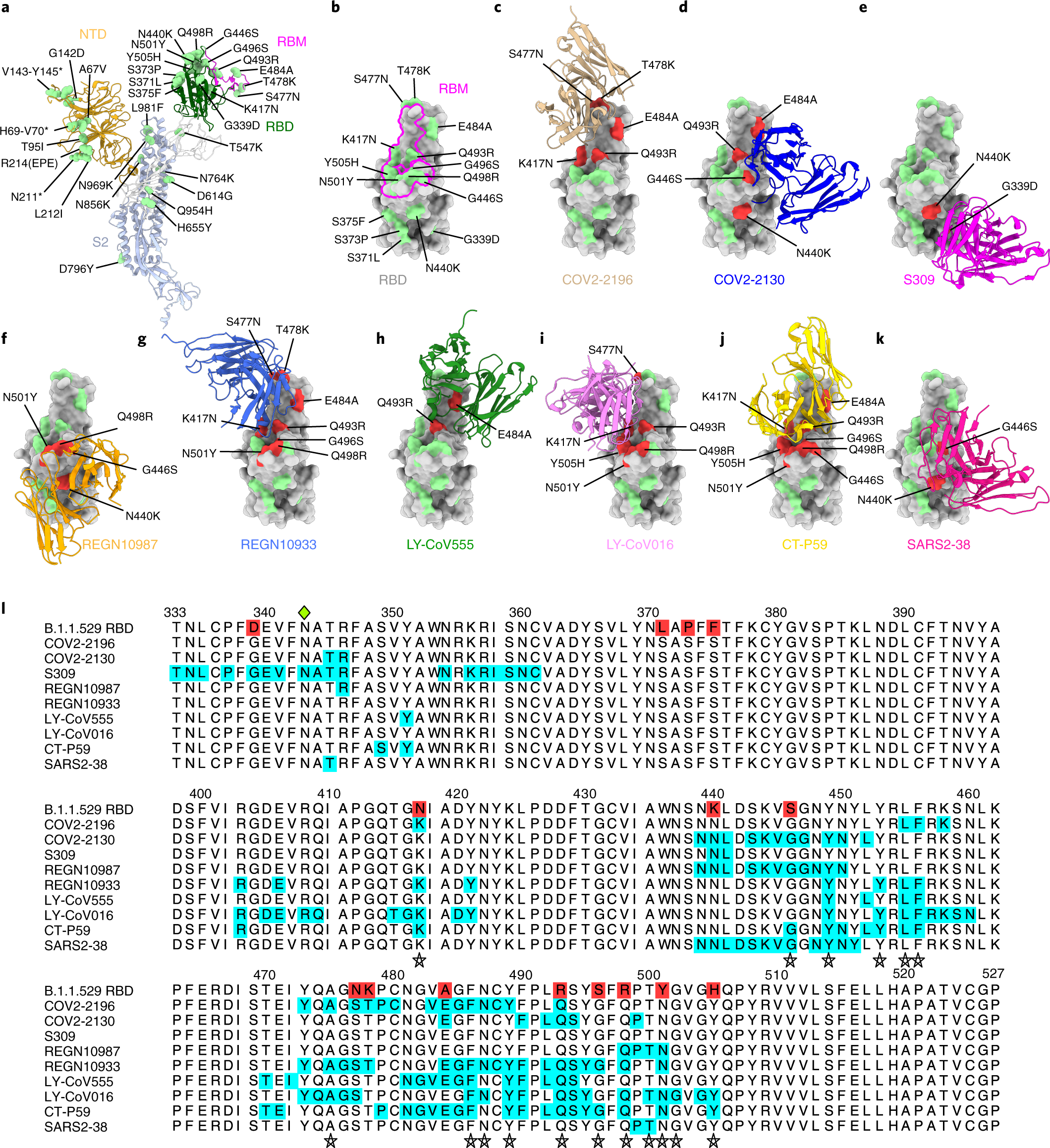
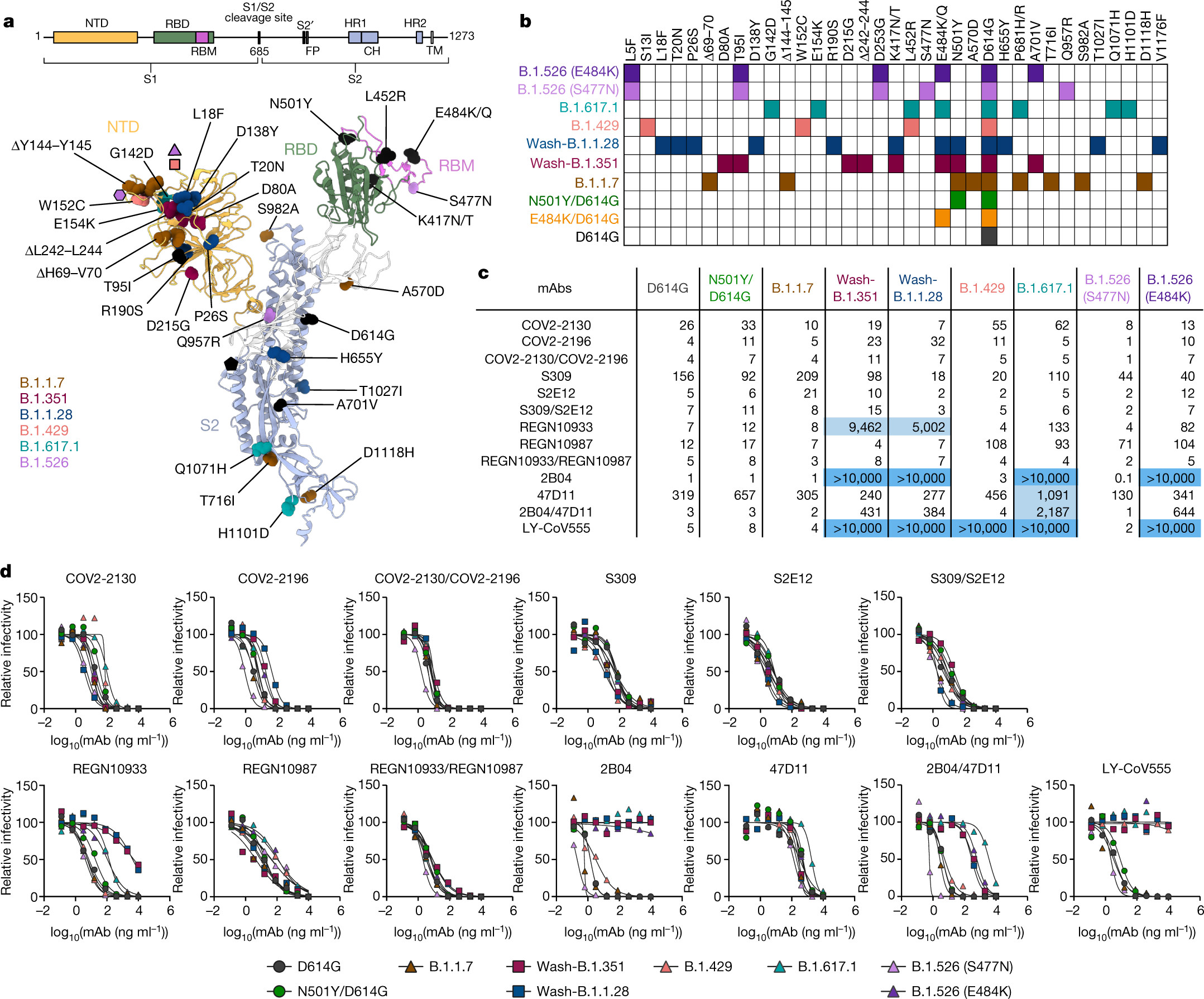
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies | Nature Medicine

Self-amplifying mRNA SARS-CoV-2 vaccines raise cross-reactive immune response to variants and prevent infection in animal models: Molecular Therapy - Methods & Clinical Development

Multivalent S2-based vaccines provide broad protection against SARS-CoV-2 variants of concern and pangolin coronaviruses - eBioMedicine
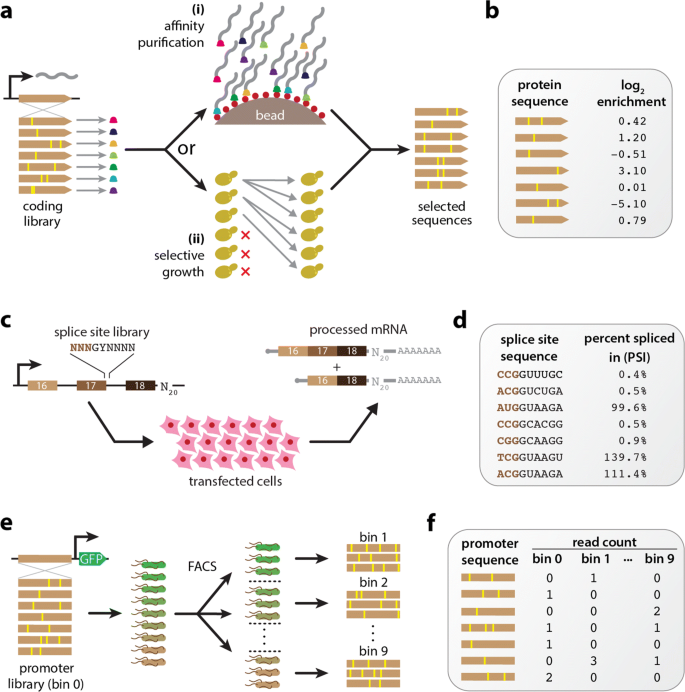
MAVE-NN: learning genotype-phenotype maps from multiplex assays of variant effect | Genome Biology | Full Text

Experimental Evidence for Enhanced Receptor Binding by Rapidly Spreading SARS-CoV-2 Variants - ScienceDirect

In vivo characterization of emerging SARS-CoV-2 variant infectivity and human antibody escape potential - ScienceDirect
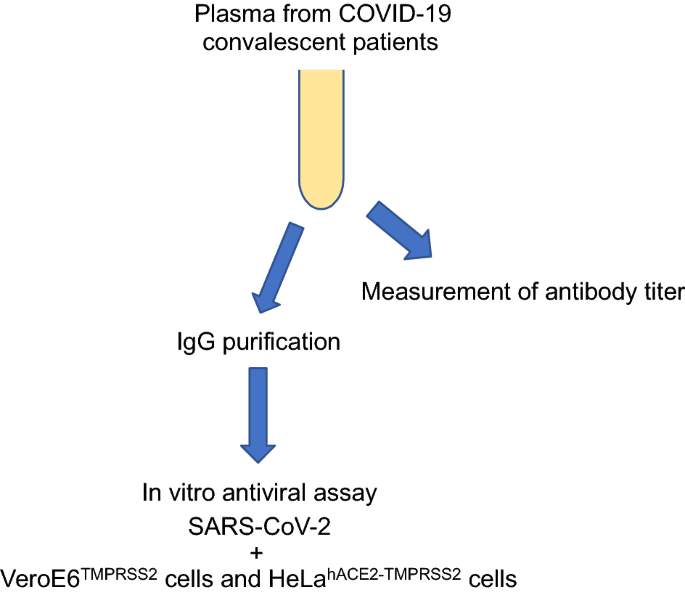
Neutralization activity of IgG antibody in COVID‑19‑convalescent plasma against SARS-CoV-2 variants | Scientific Reports

Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants | Nature Communications

SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex | Science

Combining an optimized mRNA template with a double purification process allows strong expression of in vitro transcribed mRNA - ScienceDirect